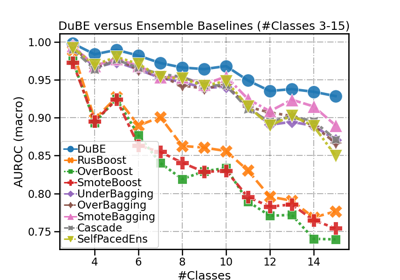
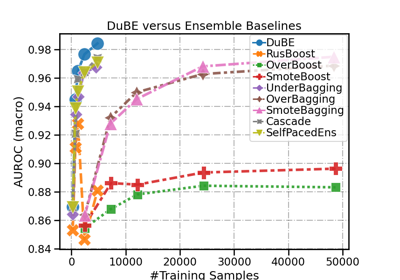
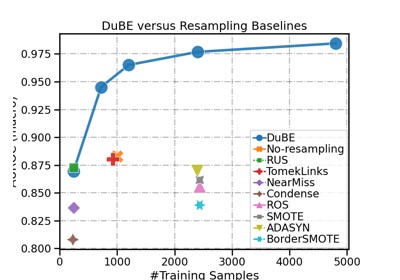
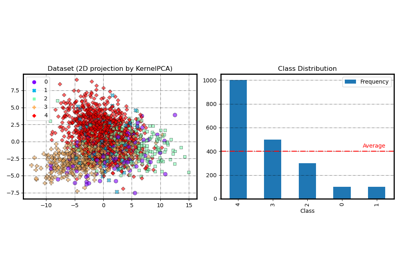
DupleBalanceClassifier
- class duplebalance.DupleBalanceClassifier(base_estimator=None, n_estimators: int = 50, perturb_alpha: float = 0.0, k_bins: int = 5, estimator_params=(), n_jobs=None, random_state=None, verbose=0)
Duple-balanced Ensemble (DuBE) Classifier for class-imbalanced learning.
DuBE is an ensemble learning framework for multi-class imbalanced classification. It simultaneously performs inter-class and intra-class balancing during the ensemble training. It is an easy-to-use solution for class-imbalanced problems, features outstanding computing efficiency, good performance, and wide compatibility with different learning models.
- Parameters
- base_estimatorestimator object, default=None
The base estimator to fit on self-paced under-sampled subsets of the dataset. Support for sample weighting is NOT required, but need proper
classes_andn_classes_attributes. IfNone, then the base estimator isDecisionTreeClassifier().- n_estimatorsint, default=10
The number of base estimators in the ensemble.
- perturb_alphafloat or str, default=”auto”
The multiplier of the calibrated Gaussian noise that was add on the sampled data. It determines the intensity of the perturbation-based augmentation. If ‘auto’, perturb_alpha will be automatically tuned using a subset of the given training data.
- k_binsint, default=5
The number of error bins that were used to approximate error distribution. It is recommended to set it to 5. One can try a larger value when the smallest class in the data set has a sufficient number (say, > 1000) of samples.
- estimator_paramslist of str, default=tuple()
The list of attributes to use as parameters when instantiating a new base estimator. If none are given, default parameters are used.
- n_jobsint, default=None
The number of jobs to run in parallel for
predict().Nonemeans 1 unless in ajoblib.parallel_backendcontext.-1means using all processors. See Glossary for more details.- random_stateint, RandomState instance or None, default=None
Control the randomization of the algorithm. If the base estimator accepts a random_state attribute, a different seed is generated for each instance in the ensemble. Pass an
intfor reproducible output across multiple function calls.If
int,random_stateis the seed used by the random number generator;If
RandomStateinstance, random_state is the random number generator;If
None, the random number generator is theRandomStateinstance used bynp.random.
- verboseint, default=0
Controls the verbosity when predicting.
- Attributes
- base_estimatorestimator
The base estimator from which the ensemble is grown.
- base_sampler_DupleBalanceHybridSampler
The base sampler.
- estimators_list of estimator
The collection of fitted base estimators.
- samplers_list of DupleBalanceHybridSampler
The collection of fitted samplers.
- classes_ndarray of shape (n_classes,)
The classes labels.
- n_classes_int
The number of classes.
- estimators_n_training_samples_list of ints
The number of training samples for each fitted base estimators.
Notes
See Basic usage example of DupleBalanceClassifier for an example.
Examples
>>> from duplebalance import DupleBalanceClassifier >>> from sklearn.datasets import make_classification >>> >>> X, y = make_classification(n_samples=1000, n_classes=3, ... n_informative=4, weights=[0.2, 0.3, 0.5], ... random_state=0) >>> clf = DupleBalanceClassifier(random_state=0) >>> clf.fit(X, y) DupleBalanceClassifier(...) >>> clf.predict(X) array([...])
Methods
fit(X, y, *[, sample_weight])Build a DuBE classifier from the training set (X, y).
get_params([deep])Get parameters for this estimator.
predict(X)Predict class for X.
Predict class probabilities for X.
score(X, y)Return the balanced AUROC score on the given test data and labels.
set_params(**params)Set the parameters of this estimator.
- fit(X, y, *, sample_weight=None, **kwargs)
Build a DuBE classifier from the training set (X, y).
- Parameters
- X{array-like, sparse matrix} of shape (n_samples, n_features)
The training input samples. Sparse matrix can be CSC, CSR, COO, DOK, or LIL. DOK and LIL are converted to CSR.
- yarray-like of shape (n_samples,)
The target values (class labels).
- resampling_target{‘hybrid’, ‘under’, ‘over’, ‘raw’}, default=”hybrid”
Determine the number of instances to be sampled from each class (inter-class balancing).
If
under, perform under-sampling. The class containing the fewest samples is considered the minority class \(c_{min}\). All other classes are then under-sampled until they are of the same size as \(c_{min}\).If
over, perform over-sampling. The class containing the argest number of samples is considered the majority class \(c_{maj}\). All other classes are then over-sampled until they are of the same size as \(c_{maj}\).If
hybrid, perform hybrid-sampling. All classes are under/over-sampled to the average number of instances from each class.If
raw, keep the original size of all classes when resampling.
- resampling_strategy{‘hem’, ‘shem’, ‘uniform’}, default=”shem”
Decide how to assign resampling probabilities to instances during ensemble training (intra-class balancing).
If
hem, perform hard-example mining. Assign probability with respect to instance’s latest prediction error.If
shem, perform soft hard-example mining. Assign probability by inversing the classification error density.If
uniform, assign uniform probability, i.e., random resampling.
- replacementbool, default=True
Whether samples are drawn with replacement. If False, sampling without replacement is performed (not applicable to over/hybrid-sampling).
- perturb_alphafloat or str, default=”auto”
The temporary value for perturb_alpha in a single function call. The multiplier of the calibrated Gaussian noise that was add on the sampled data. It determines the intensity of the perturbation-based augmentation. If ‘auto’, perturb_alpha will be automatically tuned using a subset of the given training data.
- k_binsint, default=5
The temporary value for k_bins in a single function call. The number of error bins that were used to approximate error distribution. It is recommended to set it to 5. One can try a larger value when the smallest class in the data set has a sufficient number (say, > 1000) of samples.
- sample_weightarray-like of shape (n_samples,), default=None
Sample weights for base classifier training. If None, the sample weights are initialized to
1 / n_samples.- eval_datasetsdict, default=None
Dataset(s) used for evaluation during the ensemble training process. The keys should be strings corresponding to evaluation datasets’ names. The values should be tuples corresponding to the input samples and target values.
Example:
eval_datasets = {'valid' : (X_valid, y_valid)}- eval_metricsdict, default=None
Metric(s) used for evaluation during the ensemble training process.
If
None, use the weighted (class-balanced) roc_auc_score by default.If
dict, the keys should be strings corresponding to evaluation metrics’ names. The values should be tuples corresponding to the metric function (callable) and additional kwargs (dict).The metric function should at least take 2 positional arguments
y_true,y_pred, and returns afloatas its score.The metric additional kwargs should specify the additional arguments that need to be passed into the metric function.
Example:
{'weighted_f1': (sklearn.metrics.f1_score, {'average': 'weighted'})}- train_verbosebool, int or dict, default=True
Controls the verbosity during ensemble training/fitting.
If
bool:Falsemeans disable training verbose.Truemeans print training information to sys.stdout use default setting:'granularity':int(n_estimators/10)'print_distribution':True'print_metrics':True
If
int, print information pertrain_verboserounds.If
dict, control the detailed training verbose settings. They are:'granularity': corresponding value should beint, the training information will be printed pergranularityrounds.'print_distribution': corresponding value should bebool, whether to print the data class distribution after resampling. Will be ignored if the ensemble training does not perform resampling.'print_metrics': corresponding value should bebool, whether to print the latest performance score. The performance will be evaluated on the training data and all given evaluation datasets with the specified metrics.
Warning
Setting a small
'granularity'value with'print_metrics'enabled can be costly when the training/evaluation data is large or the metric scores are hard to compute. Normally, one can set'granularity'ton_estimators/10.
- Returns
- selfobject
- get_params(deep=True)
Get parameters for this estimator.
- Parameters
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns
- paramsdict
Parameter names mapped to their values.
- predict(X)
Predict class for X.
The predicted class of an input sample is computed as the class with the highest mean predicted probability. If base estimators do not implement a
predict_probamethod, then it resorts to voting.- Parameters
- X{array-like, sparse matrix} of shape (n_samples, n_features)
The training input samples. Sparse matrices are accepted only if they are supported by the base estimator.
- Returns
- yndarray of shape (n_samples,)
The predicted classes.
- predict_proba(X)
Predict class probabilities for X.
The predicted class probabilities of an input sample is computed as the mean predicted class probabilities of the base estimators in the ensemble. If base estimators do not implement a
predict_probamethod, then it resorts to voting and the predicted class probabilities of an input sample represents the proportion of estimators predicting each class.- Parameters
- X{array-like, sparse matrix} of shape = [n_samples, n_features]
The training input samples. Sparse matrices are accepted only if they are supported by the base estimator.
- Returns
- parray of shape = [n_samples, n_classes]
The class probabilities of the input samples.
- score(X, y)
Return the balanced AUROC score on the given test data and labels. In multi-label imbalanced classification, this is an unbiased metric. The roc_auc score is calculated for each label, and find their average, weighted by support (the number of true instances for each label).
- Parameters
- Xarray-like of shape (n_samples, n_features)
Test samples.
- yarray-like of shape (n_samples,) or (n_samples, n_outputs)
True labels for X.
- sample_weightarray-like of shape (n_samples,), default=None
Sample weights.
- Returns
- ——-
- scorefloat
Balanced AUROC score of
self.predict_proba(X)wrt. y.
- set_params(**params)
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters
- **paramsdict
Estimator parameters.
- Returns
- selfestimator instance
Estimator instance.